|
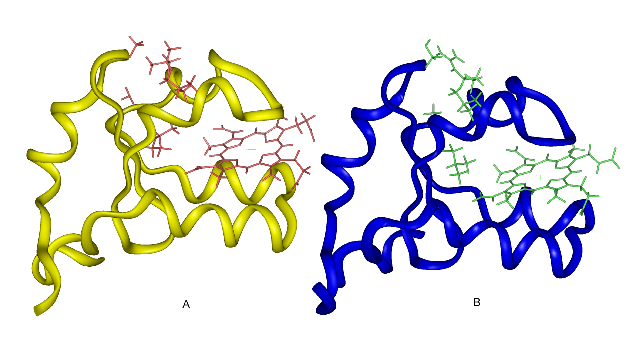
| Crystal structures of rat OM (A) and bovine Mc (B) cyt b5. At top: cleft with residues 18 and 47 in stick models. |
_______________________________________________________________________________
K.-H. Lee and K. Kuczera
Biopolymers, 69:260-269 (2003).
Two forms of cytochrome b5 have been identified, associated with the outer membrane of liver mitochondria (OM cyt b5) and with the membrane of the endoplasmic reticulum (microsomal, or Mc cyt b5). These proteins have very similar structures, but differ significantly in physical properties, with the OM cyt b5 exhibiting a more negative reduction potential, higher stability and stronger interactions with the heme. We perform molecular dynamics simulations to probe the structures and fluctuations of the two proteins in solution, to help explain the observed physical differences. We find that the structures of the two proteins, highly similar in the crystal, differ in position of a surface loop involving residues 49-51 in solution. Hydrophobic residues Ala-18, Ile-32, Leu-36 and Leu-47 tend to cluster together on the surface of rat OM cyt b5, blocking water access to the protein interior. In bovine Mc cyt b5, two of these positions, Ser-18 and Arg-47, are occupied by hydrophilic residues. This leads to breaking the hydrophobic cluster and allowing the protein to occupy a more open conformation. A measure of this structural transition is the opening of a cleft on the protein surface, which is 5 A wider in the OM cyt b5 simulation compared to the Mc form. The OM protein also appears to have a more compact hydrophobic core in its beta-sheet region. These effects may be used to explain observed stability differences between the two proteins.

|
| Crystal structures of rat OM (A) and bovine Mc (B) cyt b5. At top: cleft with residues 18 and 47 in stick models. |
_______________________________________________________________________________