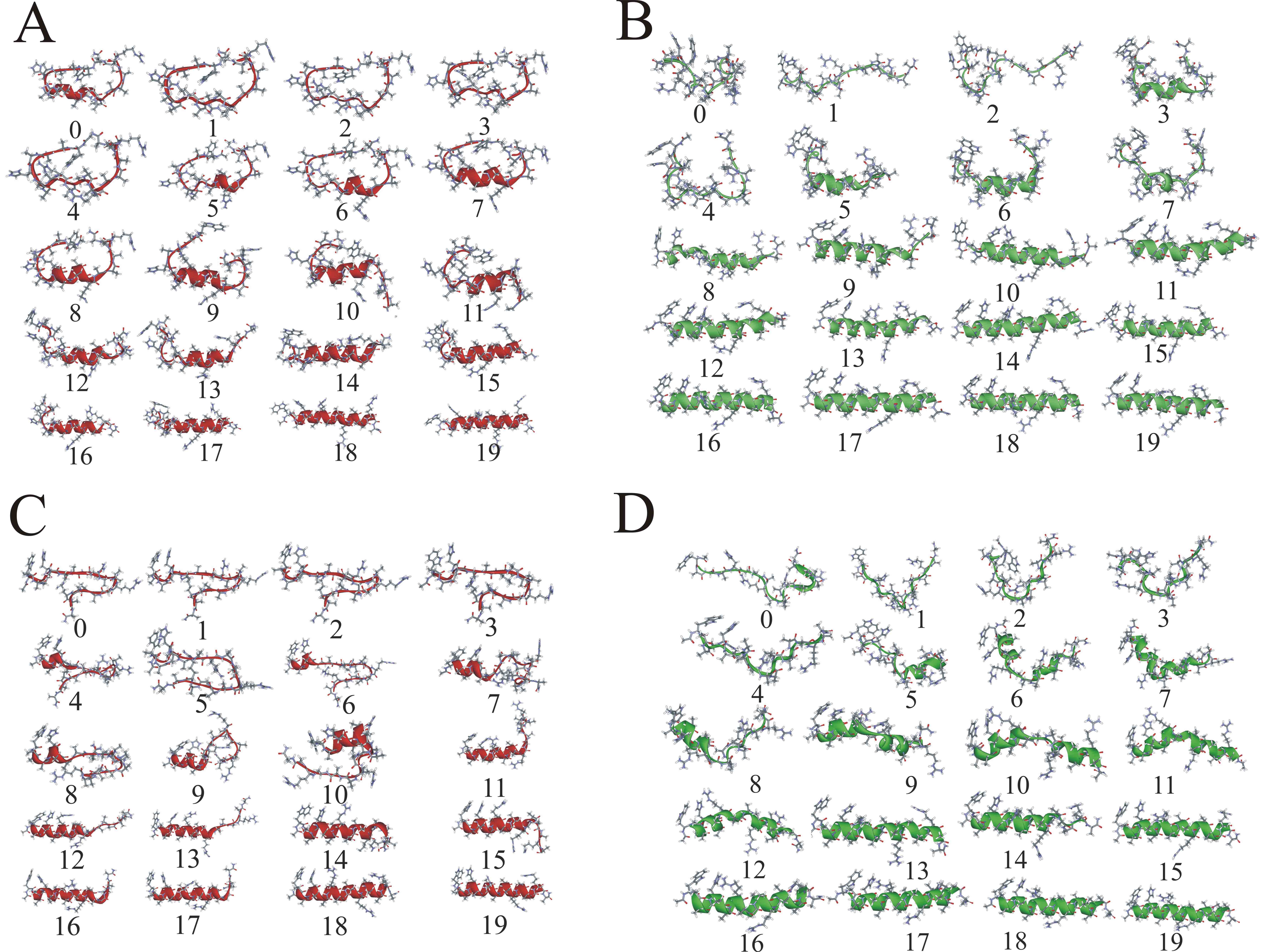
| Four main folding paths of the model WH21 helix. |
_______________________________________________________________________________
N. Ploscariu, K. Kuczera, K.E. Malek, M. Wawrzyniuk, A. Dey and R. Szoszkiewicz
J. Phys. Chem. B 118:4761-4770 (2014)
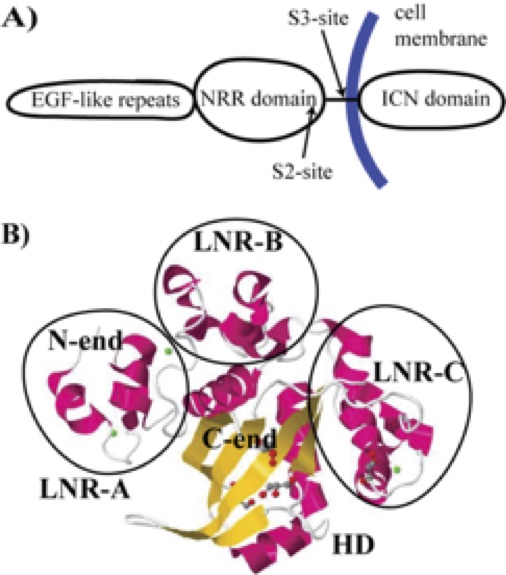
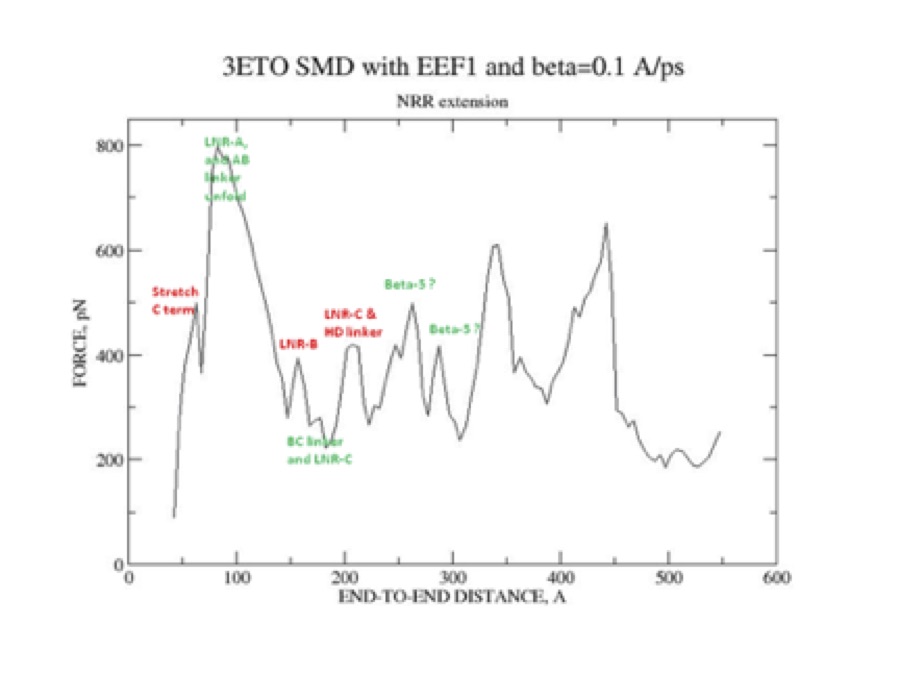
Notch signalling in metazoans is responsible for key cellular processes related to embryonic development and tissue homeostasis. Protealitic cleavage of the S2 site within an extracellular NRR domain of Notch is a key early event in Notch signaling. We use single molecule force-extension (FX) AFM force spectroscopy to investigate the role of mechanical force in unfolding the NRR domain from human Notch 1, and exposure of the S2 site. Our FX AFM measurements provide a histogram of the N to C termini lengths related to conformational transitions within the NRR domain. We detect four classes of such conformational transitions, and report that molecular forces involved in the Notch activation process are of the order of tens of pN. Based on our steered molecular dynamics (SMD) results, we associate first three classes of such events with the S2 site exposure, and obtain that mean unfolding forces for those classes are 69 +- 42 pN, 79 +- 45 pN, 90 +- 50 pN, respectively. We also provide a conditional probability analysis of the AFM data to support the hypothesis that a whole sequence of conformational transition within those three clases is the most probable pathway for the force-induced S2 site exposure. Consequently, our results provide a direct evidence at the single molecule level that a force-induced Notch activation process requires a ligand binding, so that to exert mechanical force not in random, but in several strokes and over a substantial period of time.
|
|
| Structure of Notch NRR domain. | Steered MD simulations of notch pulling. |
_______________________________________________________________________________
Jas, W.A. Hegefeld, C.R Middaugh, C.K. Johnson and K. Kuczera
J. Phys. Chem B 118:7233-7246 (2014)
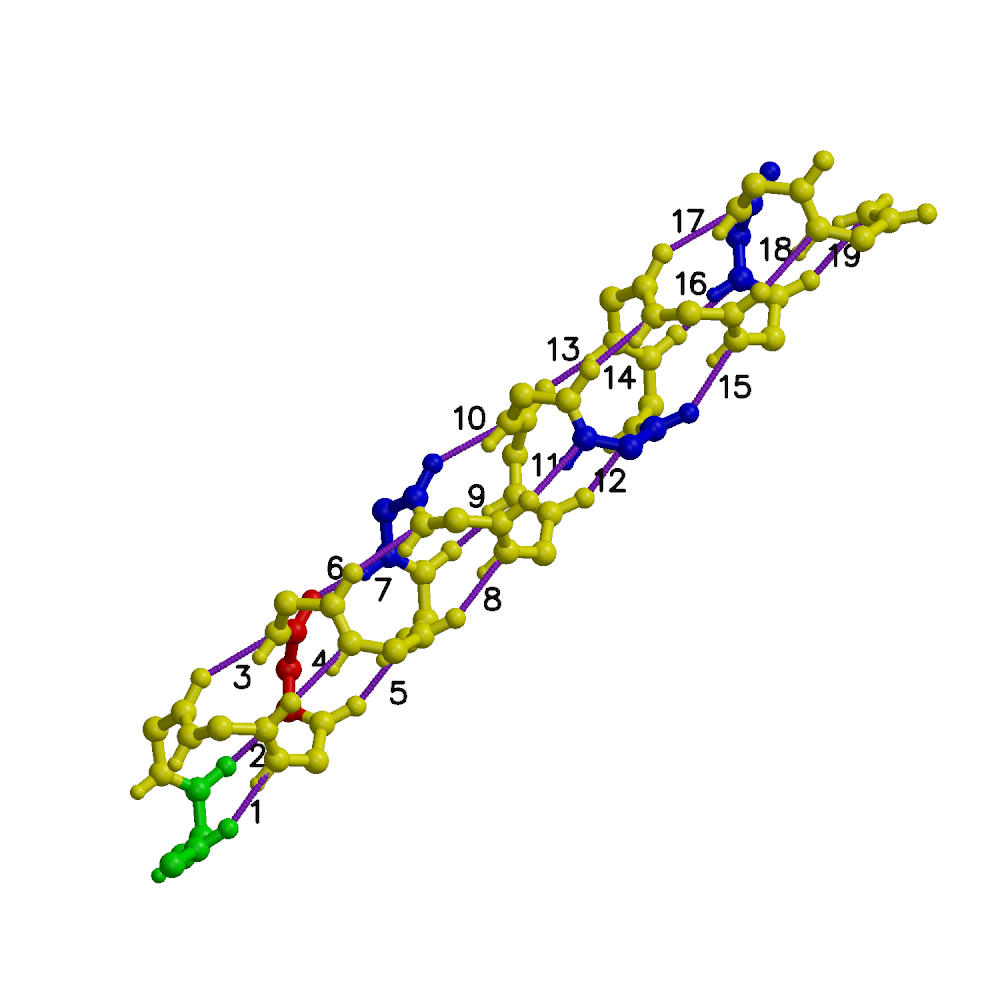
We present a combined experimental and computational study of unfolding pathways of a model 21-residue alpha-helical heteropeptide (W1H5-21) and a 16-residue beta-hairpin (GB41-56). Experimentally, we measured fluorescence energy transfer efficiency as a function of temperature, employing natural tryptophans as donors and dansylated lysines as acceptors. Secondary structural analysis was performed with circular dichroism and Fourier Transform Infrared spectroscopy. Our studies present markedly different unfolding pathways of the two elementary secondary structural elements. During thermal denaturation the helical peptide exhibits an initial decrease in length, followed by an increase, while the hairpin undergoes a systematic increase in length. In the complementary computational part of the project we performed microsecond length replica-exchange molecular dynamics simulations of the peptides in explicit solvent, yielding a detailed microscopic picture of the unfolding processes. For the helical peptide, we found a large heterogeneous population of intermediates that are primarily frayed single helices or helix-turn-helix motifs. Unfolding starts at the termini and proceeds through a stable helical region in the interior of the peptide, but shifted off-center toward the C-terminus. The simulations explain the experimentally observed non-monotonic variation of helix length with temperature as due primarily to the presence of frayed-end single-helix intermediate structures. For the hairpin peptide our simulations indicate that folding is initiated at the turn, followed by formation of the hairpin in zipper-like fashion, with CA...CA contacts propagating from the turn to termini, and hairpin hydrogen bonds forming in parallel with these contacts. In the early stages of hairpin formation the hydrophobic side-chain contacts are only partly populated. Intermediate structures with low numbers of beta-hairpin hydrogen bonds have very low populations. This is in accord with the 'broken zipper' model of Scheraga. The monotonic increase in length with temperature may be explained by the zipper-like breaking of the hairpin hydrogen bonds and backbone contacts.
|
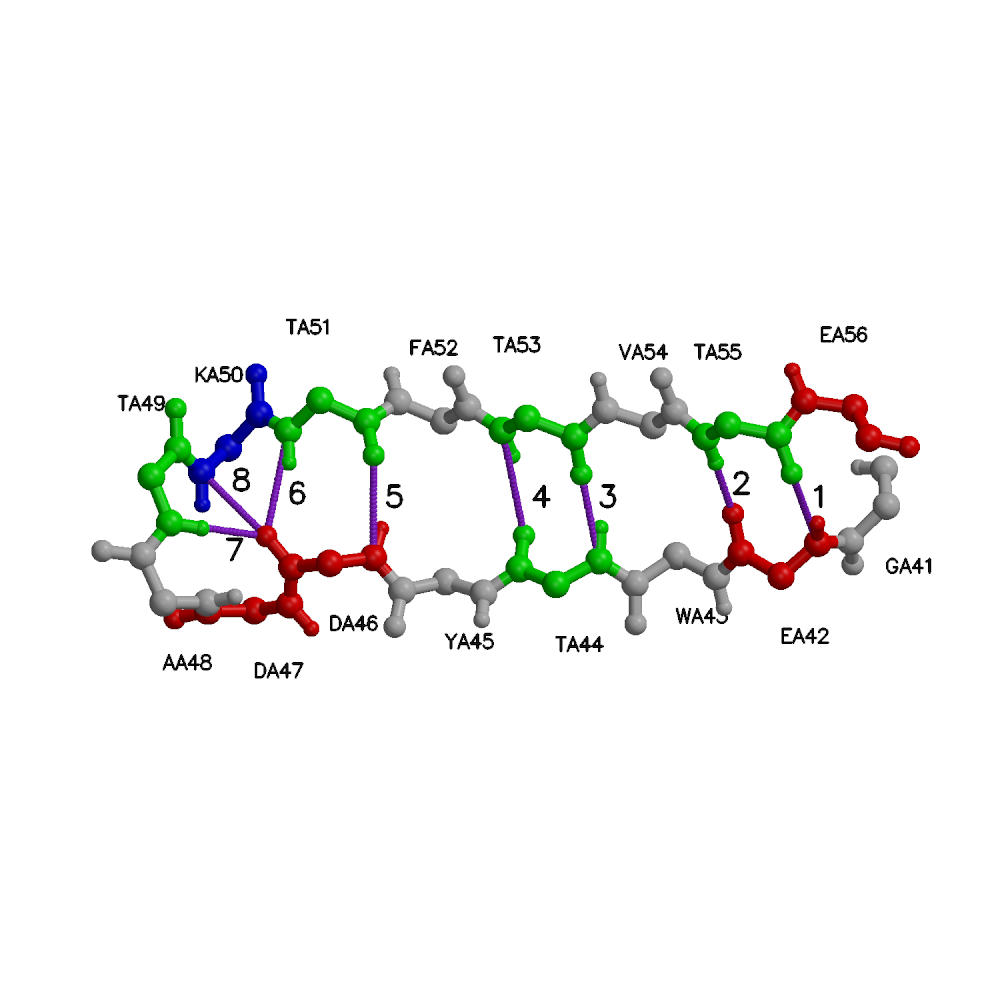
|
| Folded W1H5-21 with hydrogen bonds. | Folded GB1 with hydrogen bonds. |